Research presents new method to map cells
Apr 29, 2024
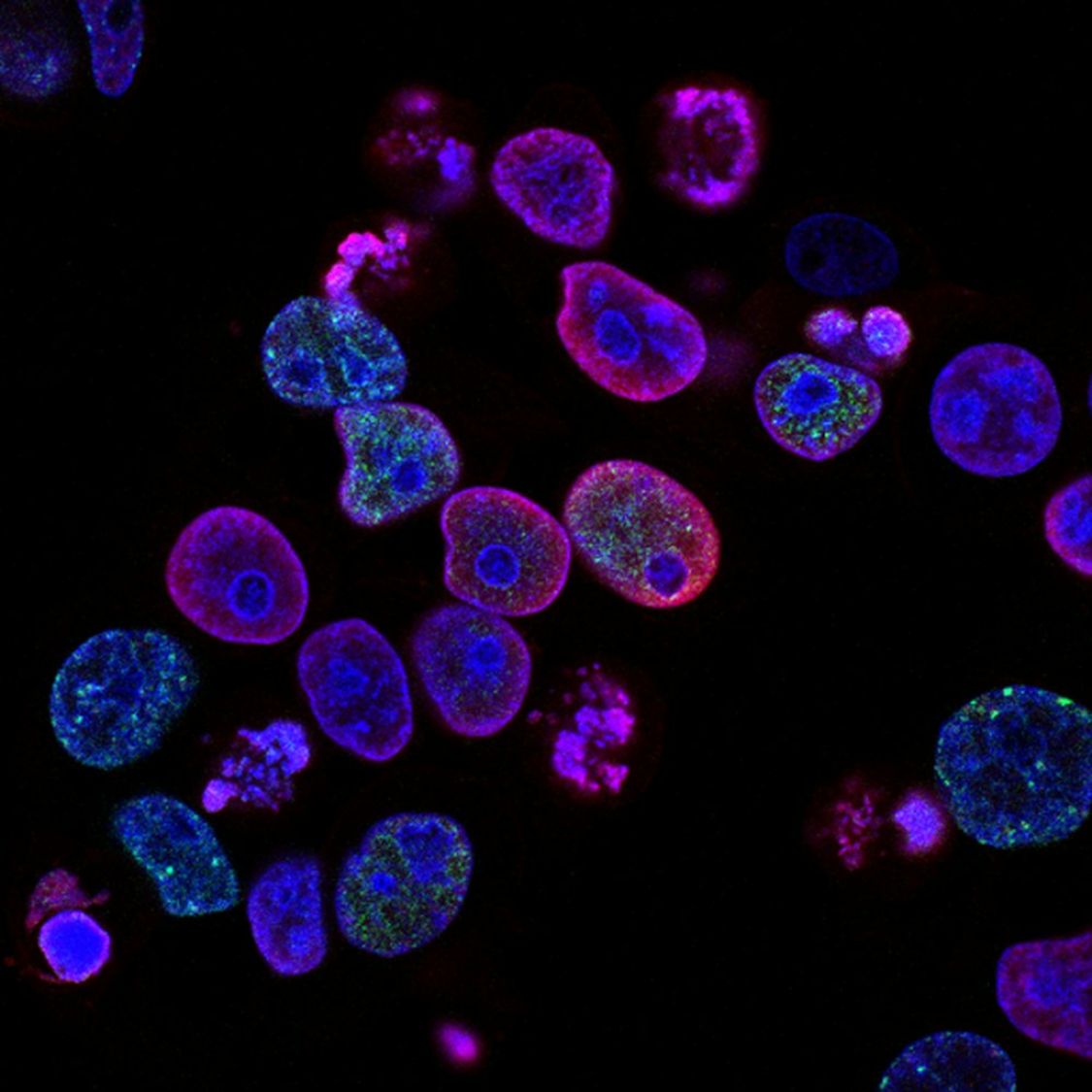
Peking University, April 29, 2024: Due to the high transparency of cells, it is very difficult to observe the organelles within them. Biologists can label specific organelles for observation through fluorescence staining. This is somewhat analogous to being in an environment without light where everyone is entirely black, making it very difficult to find your friends. By having our friends hold a fluorescent stick, we can easily locate them. An interesting question is: if the angle of the fluorescent stick held by my friend represents a kind of signal, how can we detect such angular information?
Just like this puzzle, due to the highly transparent nature of cells, it is very difficult to observe the organelles in them. With fluorescent staining, biologists can label specific organelles for observation. Most fluorescent molecules appear as directional dipoles during absorption or emission. The orientation of fluorophores can reveal important information about the structure and dynamics of their associated organelles. Fluorescence polarization microscopy has also developed as an indispensable tool for studying the orientation characteristics of biomolecules.
To overcome the challenge of conventional fluorescence polarization microscopy limited by optical diffraction, improved super-resolution fluorescence polarization microscopy techniques have been proposed, such as single-molecule orientation-localization microscopy (SMOLM) and polarization modulation (e.g., SDOM, SPoD, etc.). However, from the biotechnological point of view, despite the significant role of biological filaments (e.g., actin filaments and microtubules) in cellular functions, there is a lack of approaches with 3D orientation resolving and high temporal-spatial resolution to study them in vivo.
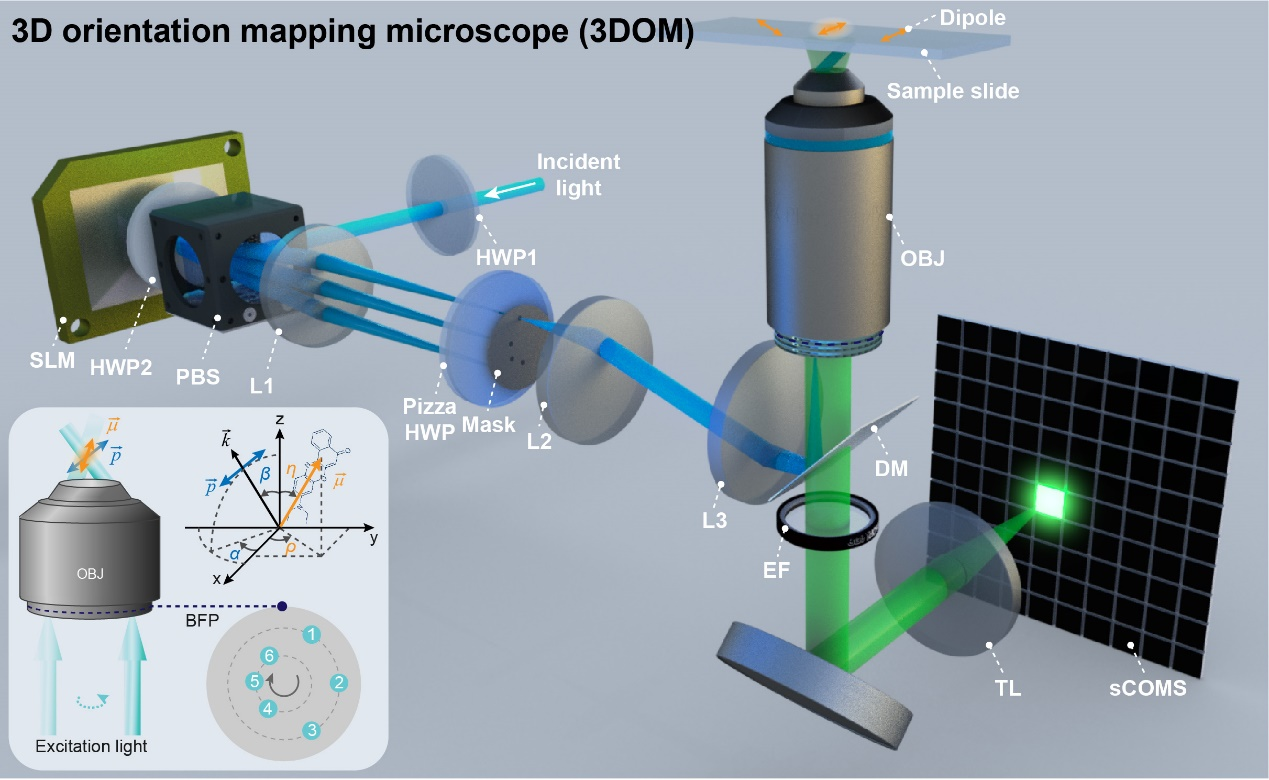
Figure 1: Schematic illustration of the 3DOM microscopy.
To address the problem of dipole orientation resolution, Professor Xi Peng's research group from Peking University has developed a 2D dipole orientation mapping method, SDOM (Light.: Sci. Appl., 2016), and optical lock-in detection super-resolution dipole orientation mapping, OLID-SDOM (Light.: Sci. Appl., 2022). In a recent issue of PhotoniX, the research group reported a super-resolution 3D orientation mapping microscope termed 3DOM.
The 3DOM method is based on the polarized structured illumination microscopy developed by the research group. Reversing the principle of Young's double-slit interference and combining it with the principle of reversible light paths, different angles of the stripes are used to produce positive and negative first-order beams in different directions. Furthermore, a single direction of tilted illumination can be produced by simply blocking the corresponding negative first-order light. By projecting this tilt to different angles of the z-axis and reconstructing the image using the FISTA algorithm, high-precision resolution of the dipole orientation can be achieved by combining the polarization modulation coefficients and the reconstruction results in reciprocal space.
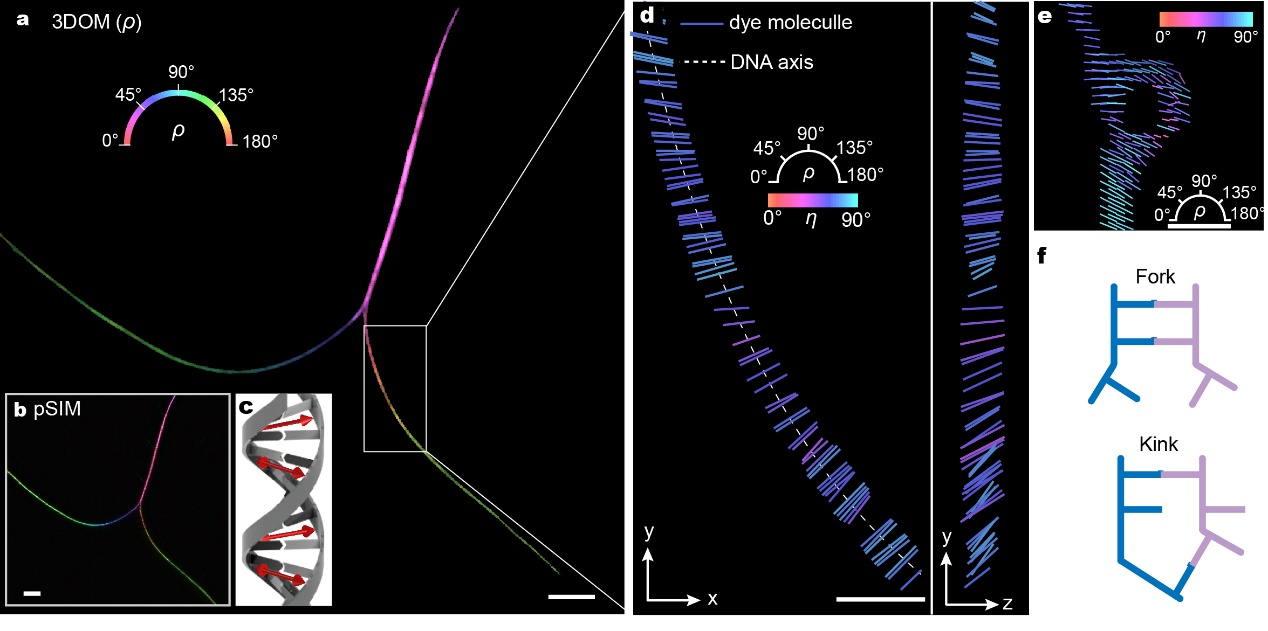 Figure 2
Overall, our proposed 3DOM method effectively overcomes the limitations of fluorescence polarization microscopy in spatial resolution and 3D orientation mapping using widefield imaging. 3DOM provides a more comprehensive understanding of the 3D spatial structure of fluorophore molecules. This enables us not only to distinguish various cytoskeletal organizations (actin filaments and microtubules) but also to gain valuable insights into filament binding compactness and the order of subcellular structures. Moreover, 3DOM holds significant potential in DNA bending and the orientation of membranous organelles. One of the key advantages of 3DOM is its ease of upgradability to existing widefield systems. The simple implementation, accurate 3D dipole orientation information, and superior spatiotemporal resolution of 3DOM make it suitable for a wide range of applications, enhancing its accessibility and usability in different research settings. This powerful tool empowers researchers to unravel the intricate complexities of subcellular structure, biomechanics, and biodynamics, revolutionizing our understanding of cellular processes. We foresee 3DOM advancing understanding across a multitude of biological structures and interactions operative at the nanoscale.
In this work, Ph.D. student Zhong Suyi from Peking University is the first author, Professor Xi Peng and Dr. Li Meiqi are the co-corresponding authors. This study received support from the National Key Research & Development Program and National Natural Science Foundation of China.
Xi Peng's research group is committed to developing new types of super-resolution microscopy technology. The work carried out in the field of polarization super-resolution is as follows: 1) Achieved super-resolution of 50nm and measured the two-dimensional dipole of fluorescence by utilizing polarization dipoles (Light: Science and Applications 2016); 2) Realized polarization structured illumination microscopy (SIM) super-resolution by resolving the polarization information in SIM, with successful commercialization (Nature Communications 2019); 3) Successfully observed and studied the environment of 10 types of organelles simultaneously by combining the chemical polarity (spectral redshift) and physical orderliness (dipole freedom) information of fluorescent molecules (Nature Communications 2020); 4) Enhanced the weak polarization modulation signal through polarization modulation and optical lock-in amplification technology, achieving high sensitivity measurement of fluorescence anisotropy of subcellular structures in living cells (Light: Science and Applications 2022); 5) Developed open-source software Open-3DSIM with dipole orientation analysis capability for 3D-SIM (Nature Methods 2023) and open-source hardware (Advanced Photonics Nexus 2024); 6) Conducted a systematic review of the relevant SIM reconstruction algorithms (Light: Science and Applications 2023).
Link: https://photonix.springeropen.com/articles/10.1186/s43074-024-00127-6
Figure 2
Overall, our proposed 3DOM method effectively overcomes the limitations of fluorescence polarization microscopy in spatial resolution and 3D orientation mapping using widefield imaging. 3DOM provides a more comprehensive understanding of the 3D spatial structure of fluorophore molecules. This enables us not only to distinguish various cytoskeletal organizations (actin filaments and microtubules) but also to gain valuable insights into filament binding compactness and the order of subcellular structures. Moreover, 3DOM holds significant potential in DNA bending and the orientation of membranous organelles. One of the key advantages of 3DOM is its ease of upgradability to existing widefield systems. The simple implementation, accurate 3D dipole orientation information, and superior spatiotemporal resolution of 3DOM make it suitable for a wide range of applications, enhancing its accessibility and usability in different research settings. This powerful tool empowers researchers to unravel the intricate complexities of subcellular structure, biomechanics, and biodynamics, revolutionizing our understanding of cellular processes. We foresee 3DOM advancing understanding across a multitude of biological structures and interactions operative at the nanoscale.
In this work, Ph.D. student Zhong Suyi from Peking University is the first author, Professor Xi Peng and Dr. Li Meiqi are the co-corresponding authors. This study received support from the National Key Research & Development Program and National Natural Science Foundation of China.
Xi Peng's research group is committed to developing new types of super-resolution microscopy technology. The work carried out in the field of polarization super-resolution is as follows: 1) Achieved super-resolution of 50nm and measured the two-dimensional dipole of fluorescence by utilizing polarization dipoles (Light: Science and Applications 2016); 2) Realized polarization structured illumination microscopy (SIM) super-resolution by resolving the polarization information in SIM, with successful commercialization (Nature Communications 2019); 3) Successfully observed and studied the environment of 10 types of organelles simultaneously by combining the chemical polarity (spectral redshift) and physical orderliness (dipole freedom) information of fluorescent molecules (Nature Communications 2020); 4) Enhanced the weak polarization modulation signal through polarization modulation and optical lock-in amplification technology, achieving high sensitivity measurement of fluorescence anisotropy of subcellular structures in living cells (Light: Science and Applications 2022); 5) Developed open-source software Open-3DSIM with dipole orientation analysis capability for 3D-SIM (Nature Methods 2023) and open-source hardware (Advanced Photonics Nexus 2024); 6) Conducted a systematic review of the relevant SIM reconstruction algorithms (Light: Science and Applications 2023).
Link: https://photonix.springeropen.com/articles/10.1186/s43074-024-00127-6
Source:
College of Future Technology